Research Interests
***Undergraduates in Research***
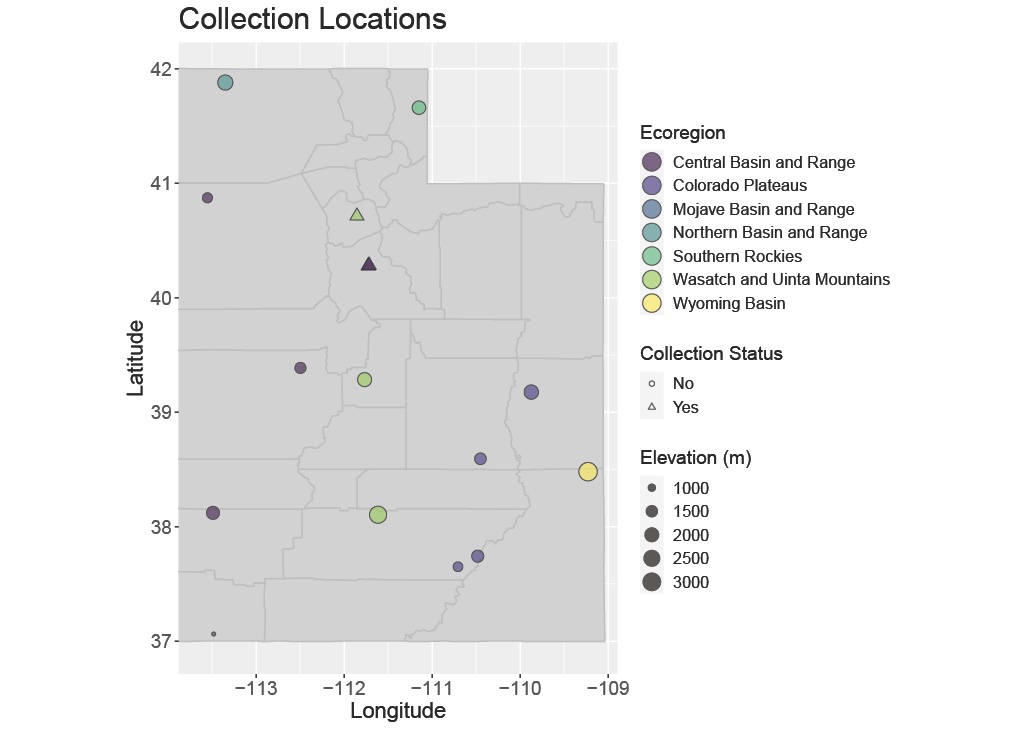
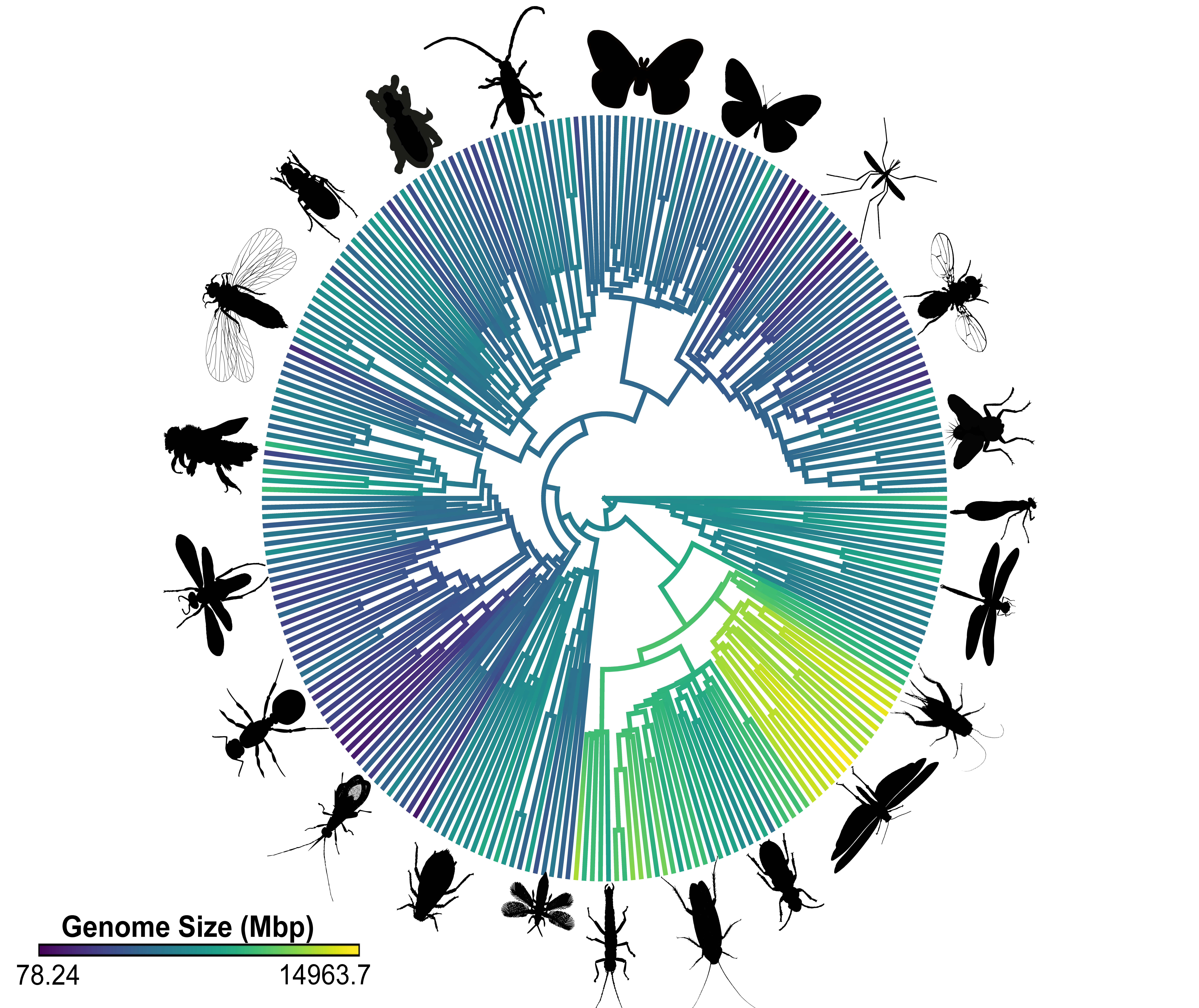
Genome Size Evolution Among Species
Genome Size Evolution Within Species
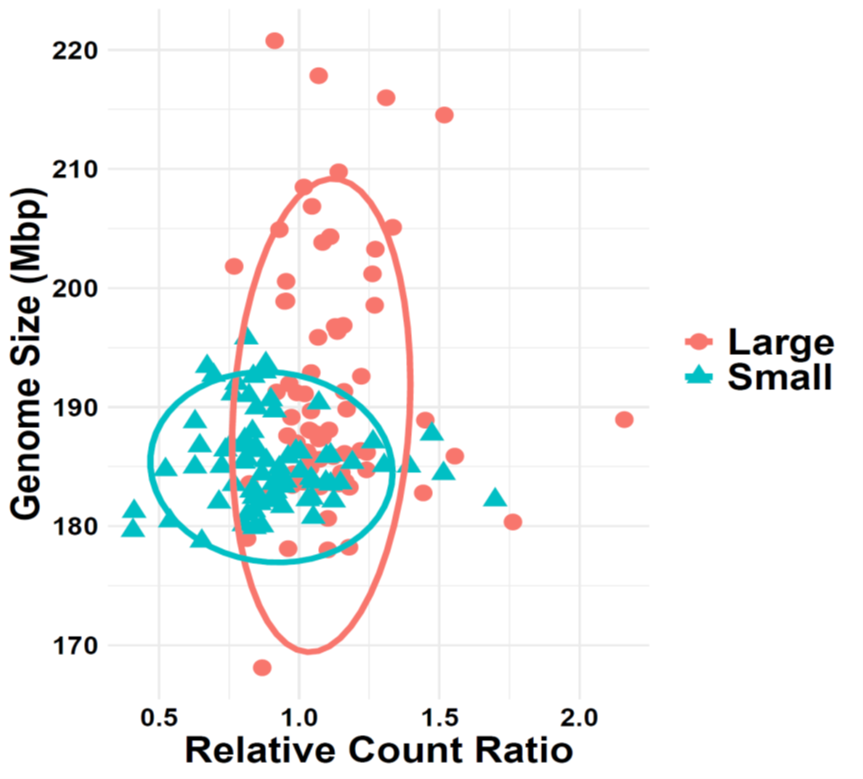
While genome size has been known to vary from species to species, genome size, like any other trait, can vary within a species. In fact, there was found to be about 30,000,000 more base pairs of DNA in the Drosophila DGRP with the largest genomes than the ones with the smallest! This within species variation has been shown to be correlated to other phenotypes, such as reproductive fitness, body size, and development time. I have utilized long running phenotype selection lines to evaluate genome size change in relatively short periods of time (Hjelmen et al 2020). I am working to investigate the evolutionary advantages (or disadvantages) to having more or less DNA. Does certain types of DNA assist in adaptation to new environments? What amount of variation in genome size is expected within a species? What contributes to this variation? And at what point does it lead to divergence/speciation?
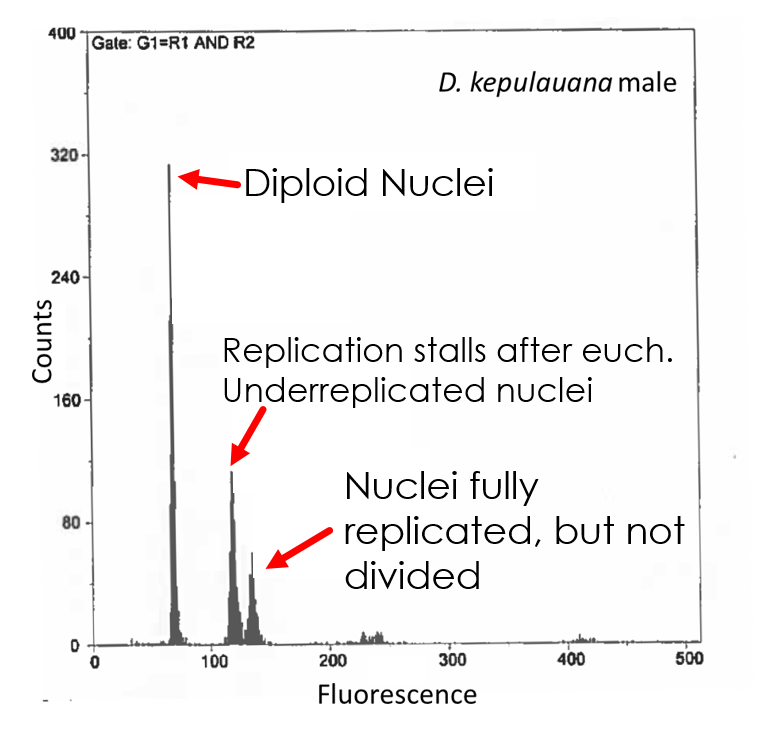
Underreplication and Heterochromatin Content of the Genome
Underreplication is a fascinating phenomenon by which DNA replication is stalled before replicating the late replicating heterochromatin. This phenomenon has been noted for decades in the polytene salivary glands of Drosophila, but only recently was documented in the thoracic tissue of D. melanogaster. Recently, we showed (with the help of some expert dissection skills from an undergrad!) what thoracic tissues underreplication is occuring in (Johnston et al 2020). I also recently demonstrated underreplication in 132 species within the Drosophila genus (Hjelmen et al. 2020)! We know variation in genome size is largely due to repetetive and noncoding DNA (largely heterochromatic), and we findthat more of this "unreplicated" DNA occurs in species with larger genomes. We therefore use Underreplication to study the dynamics of Heterochromatin and Euchromatin change throughout time. We find dramatically different patterns of change between types of chromatin. Is this dependent on what group of species we look at? What is occuring in species that have significantly more heterochromatin? Why does this partial replication occur in thoracic tissue? And why in only Drosophila? Does it have something to due with adaptation to environments? What is the physiological benefit of more DNA in the thorax? I hope to answer these questions through long read sequencing, transcriptomics, and studies of phenotypes!
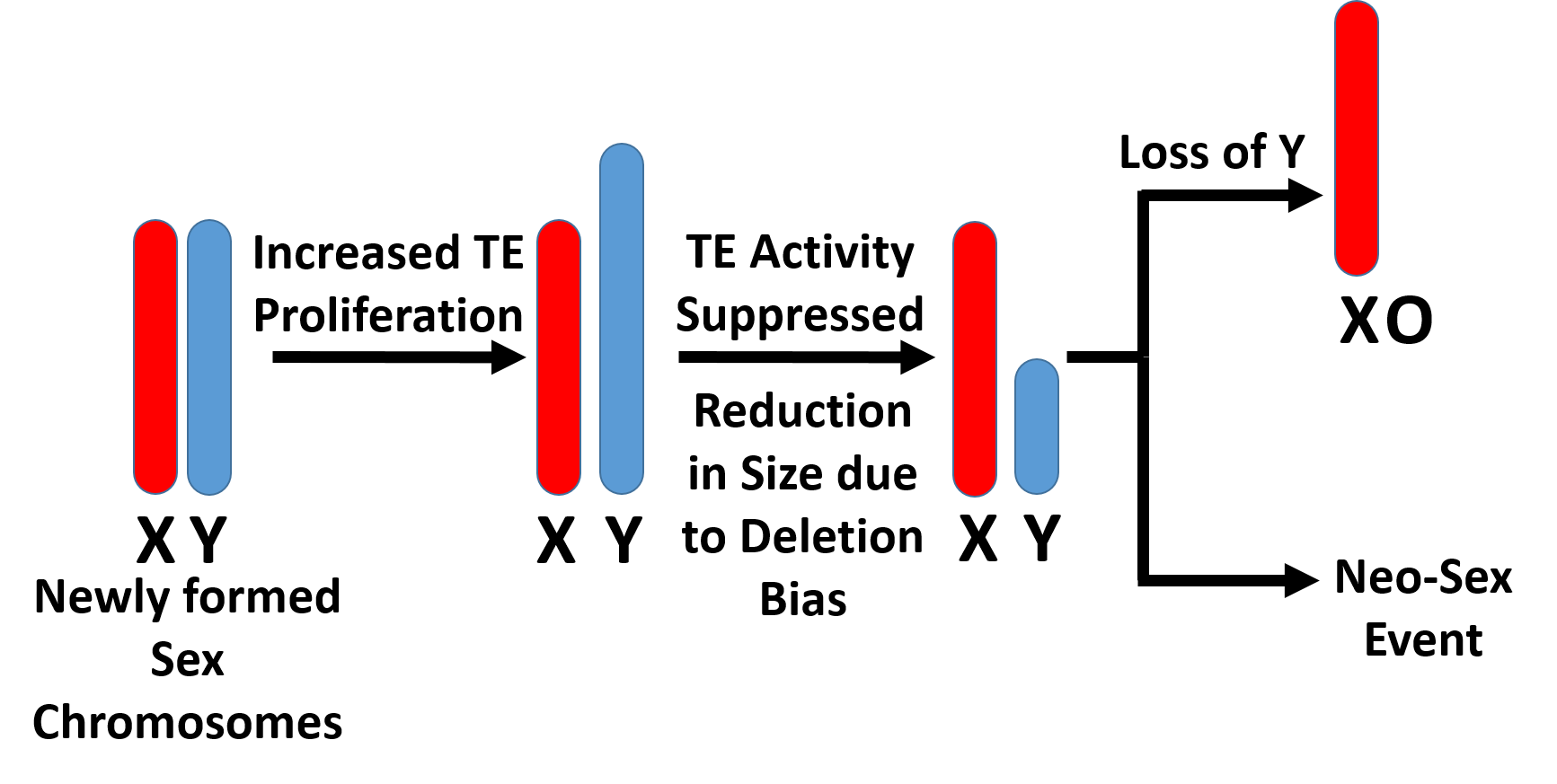
Sex Chromosome Evolution
Sex chromosomes are often the first thing to which biologists attribute differences between sexes. While most of the DNA content in a genome is on the autosomes and therefore does not differ between the sexes, differences between sex chromosomes lead to highly differentiated gene expression and phenotypes between the sexes. In the an XY sex system, as the sex chromosomes differentiate, the Y chromosome becomes more and more sparse in the case of gene content and becomes highly heterochromatized and compacted. We have found that in many cases, not only does gene content decrease, but also the physical amount of DNA on the Y chromosome decreases (Hjelmen et al 2018, Hjelmen et al 2019). Hypothetically, the Y chromosome gets smaller and smaller throughout time, until it becomes so small it can be lost. Here we may see sex chromosome turnover and introduction of Neo-sex systems. So, how big can the differences between sexes become? How often are males in XY systems larger than females? Are those with larger male genomes indicative of neo-Sex chromosomes? Beyond just looking at physical size, I am interested in seeing what content is found on neo-sex chromosomes and investigating whether or not specific chromosomes are more likely to be selected to become sex chromosomes. To do this, I am sequencing males and females from species which I have identified as having prospective sex chromosome turnover events.
Forensic Entomology
Forensic entomology is the use of insects (and related arthropods) in legal investigations. The most well known application of this work is in the medico-legal field: using insects to aid in death investigations. In order to use insects in this regard, we must have foundational information about their biology: development, occurrences, impact of abiotic, factors, etc. I have previously done work on transcriptomics of these flies (Hjelmen et al. 2021, and Pimsler et al. 2021) and other developmental applications (Hjelmen et al. 2020). We are now working to understand the distributions and occurrences of forensically relevant flies across the ecoregions of Utah. We are also working to develop other bioinformatic and genomic applications using these important flies. This is in collaboration with Dr. Lauren M. Weidner at ASU.